N armed bandit task
#n_armed_bandit.py # # n-armed bandit task chapter 2 Figure 2.1, 2.4 # # 1. implemented with softmax function or epsilon-greedy function # 2. implemented with online method for values # 3. implemented with optimistic comparison # import numpy as np import matplotlib.pyplot as plt #nBandit: the number of bandits #nArm: the number of arms #nPlay: the number of plyas (times we will pull a arm) #sigma the standard deviation of the return from each of the arms #Epsilon Greedy Method: x=qt, y=teps, z=_ def Greedy_method(Q, epsilon, numA): qt = Q[0]; if np.random.rand() <= epsilon: #epsilon part arm = np.random.randint(0, numA); else: #greedy part #arm = np.where(max(qT[bi, :])==qT[bi,:])[0][0]; arm = np.argmax(qt); return arm #Softmax method: x=qt, y=teps def Softmax_method(Q, epsilon): temperature = epsilon; qt = Q[0]; softe = np.exp(qt/temperature); #softmax function soft_max = softe / np.sum(softe); z = np.random.rand(); #cumulative probability cum_prob = 0.0; for i in xrange(len(soft_max)): prob = soft_max[i]; cum_prob += prob; if cum_prob > z: arm = i return arm return len(soft_max) - 1 #optimistic comparison #x = optomistic, y = nArm, z = ei def initial_velection(optimis, numA, te): qN = np.zeros((1, numA)); #inital values set to zeros if optimis == 0: qT = np.zeros((1, numA)); #initial values set to be optimistic else: if te == 0: qT = np.ones((1, numA)) * 5.0; #optomistic value else: qT = np.zeros((1, numA)); return qT, qN #main code #nBandit: the number of simulation #nArm: the number of Arms #nPlay: the play times #sigma: variance of rewards #func_selection: choose either epsilon-method(func_selection=1) or softmax method(others) #optimistic: set optimistically initial values(optimistic=1) or not(others) def n_armed_testbed(nBandit=2000, nArm=10, nPlay=1000, sigma=1.0, func_selection=0, optimistic=0): #function = 1 for epsilon-greedy method else for softmax function if optimistic == 0 and func_selection == 0: #A set of epsilon for softmax function eps = [0.01, 0.1, 1]; elif optimistic == 0 and func_selection == 1: #set of epsilon for greedy method eps = [0, 0.01, 0.1]; else: #comparison between optimistic or nonoptimistic case eps = [0.0, 0.1]; #the true reward from multiple gaussian distribution qTmean = np.random.multivariate_normal(np.zeros((nArm)), np.eye(nArm), (nBandit)); #initial values for [row, column] = np.shape(qTmean); qT0 = np.zeros((row,column)); #result containers average_reward = np.zeros((len(eps), nPlay)); perOptAction = np.zeros((len(eps), nPlay)); #make loops for each epsilon parameter for ei in xrange(len(eps)): #pick up a epsilon teps = eps[ei]; #initialization of Action Values #make loops for each bandit Rewards = np.zeros((nBandit, nPlay)); optAction = np.zeros((nBandit, nPlay)); for bi in xrange(nBandit): #optimistic values #qT = np.zeros((1, nArm)); #initialization of values #qN = np.zeros((1, nArm)); #tracks of the number draws on each arm qT, qN = initial_velection(optimistic, nArm, ei); #make loops for each play for p in xrange(nPlay): #choose either epsilon-greedy or softmax #epsilon-greedy if func_selection == 1 : arm = Greedy_method(qT, teps, nArm); #choose softmax method else: arm = Softmax_method(qT, teps); #extract best arm choice best_arm = np.argmax(qTmean[bi, :]); #if selected arm is equivalent to best_arm, then count. if arm == best_arm: optAction[bi, p] = 1.0; #get the reward from drawing on that arm with qTmean + gaussian(mean=0,sigma=1) reward = qTmean[bi, arm] + sigma * np.random.normal(0,1); Rewards[bi, p] = reward; #update qN, qT #qT[0, arm] = qT[0, arm] + (reward - qT[0, arm])/(qN[0, arm] + 1); qT[0, arm] = qT[0, arm] + 0.1*(reward - qT[0, arm]); #qN[0, arm] = qN[0, arm] + 1.0; #calculation of average action avg = np.mean(Rewards, 0); average_reward[ei, :] = avg.T; #calculation of optimal action PercentOptAction = np.mean(optAction, 0); perOptAction[ei, :] = PercentOptAction.T; #plotting label w.r.t epsilon plot_label = "epsilon="; list_label = []; for i in eps: list_label.append(plot_label + str(i)); #plotting for Average Reward x = np.linspace(0, nPlay, nPlay); for i in xrange(len(eps)): plt.plot(x, average_reward[i]); plt.xlabel('Plays'); plt.ylabel('Average Rewards'); plt.legend(list_label); plt.show() #plotting for Optimal Action for i in xrange(len(eps)): plt.plot(x, perOptAction[i]); plt.xlabel('Plays') plt.ylabel('Optimal Actions(%)') plt.legend(list_label); plt.show() def main(): n_armed_testbed(); if __name__ == "__main__": main()
1D Heat Conduction with a delta function
Analytic solution
#heat conduction with a delta function import numpy as np import matplotlib.pyplot as plt import time M = 100; #the number of division x = np.linspace(0, 1, M+1); N = 600; #finite series #T = [0.001, 0.004, 0.016, 0.064] #time t T = np.linspace(0,0.0001, M); F = np.ones((1, M+1)); for k in xrange(1, N): w = k * np.pi; F += 2 * np.cos(w/2) * np.cos(w* x) plt.ion() line, = plt.plot(x, F[0]); for t in T: F = np.ones((1, M+1)); for k in xrange(1, N): w = k * np.pi; F += 2 * np.cos(w/2) * np.cos(w* x) * np.e**(-w**2*t) line.set_ydata(F) time.sleep(0.1) plt.draw()
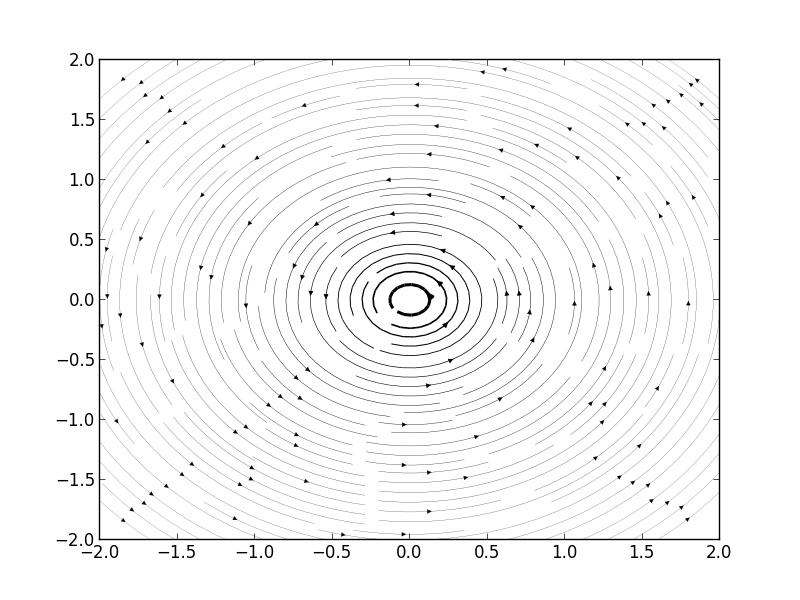
Streamlines of A Free Vortex
We think a case of
,
.
Then, when we think about velocity of this flow along x and y axis.
Then we can get Figure1.
However, when we think about inertial frame of reference.
In this particular case, velocity is equivalent to this formula.
[v_{moving} = v_{static} - v_{frame}]
Hence, we just add constant V in u.
Thus, we can get Figure 2.
import matplotlib.pyplot as plt import numpy as np x = np.linspace(-2,2,1000); y = np.linspace(-2,2,1000); C = 1; V = 1; U = 0; u = -C*y[:,np.newaxis]/(x**2 + y[:,np.newaxis]**2) - V; v = C*x/(x**2 + y[:,np.newaxis]**2) - U; speed = np.sqrt(u*u + v*v) plt.figure() plt.streamplot(x, y, u, v, density=(2,2), color='k', linewidth=100*speed/speed.max()) plt.show()
#two free vortices
import matplotlib.pyplot as plt import numpy as np x = np.linspace(-2,2,1000); y = np.linspace(-2,2,1000); C = 5; D = -1; V = 0; U = 0; x0 = 1; x1 = -1; u = -C*y[:,np.newaxis]/(2*np.pi*((x-x0)**2 + y[:,np.newaxis]**2)) - D*y[:,np.newaxis]/(2*np.pi*((x-x1)**2 + y[:,np.newaxis]**2)) - V; v = C*(x-x0)/(2*np.pi*((x-x0)**2 + y[:,np.newaxis]**2)) + D*(x-x1)/(2*np.pi*((x-x1)**2 + y[:,np.newaxis]**2)) - U; speed = np.sqrt(u*u + v*v) plt.figure() plt.streamplot(x, y, u, v, density=(2,2), color='k', linewidth=100*speed/speed.max()) plt.show()
C++ compiling with Mac terminal
gcc -c main.cpp gcc -o main.cgi main.o -lstdc++ ./main.cgi
Synaptic Conductance(alpha function, Python)
import math import numpy as np import matplotlib.pyplot as plt def trapsyn(dt, Tfin, gsyn): VCl = -68; #mV Cm = 1; #micro F/cm^2 gCl = 0.3; #mS/cm^2 z = 2./dt; Nt = int(math.ceil(1 + Tfin / dt)); #number of time steps V = np.zeros((Nt, 1)); t = np.zeros((Nt, 1)); #preallocate space g = np.zeros((Nt, np.size(gsyn['gmax']) )); j = 0; V[0] = VCl; a0 = gCl / Cm; b0 = gCl * VCl / Cm; for j in xrange(1, Nt): t[ j ] = (j - 1) * dt; g[j, :] = gsyn['gmax'] * ((t[j] -gsyn['t1']) / gsyn['taua'] ) * np.exp(1 - (t[j] - gsyn['t1'])/gsyn['taua']) * ( t[j] > gsyn['t1'] ) a1 = (gCl + sum(g[j, :])) / Cm; tmp = np.dot(g[j, :], gsyn['Vsyn'].T); b1 = (gCl*VCl + np.dot(g[j, :], gsyn['Vsyn'].T) )/Cm; V[ j ] = ( (z - a0) * V[j-1] + b0 + b1) / (z + a1); a0 = a1; b0 = b1 return t, V, g def main(): gsyn = {"gmax":np.array([[0.2]]), "taua":np.array([[2.0]]), "t1":np.array([[5.0]]), "Vsyn":np.array([[0.0]])}; [t, V1, g1] = trapsyn(0.01, 35, gsyn); gsyn2 = {"gmax":0.2 * np.array([[1., 1.]]), "taua":2 * np.array([[1., 1.]]), "t1":np.array([[4., 5.]]), "Vsyn":np.array([[-68, 0]])} [t, V2, g2] = trapsyn(0.01, 35, gsyn2); plt.plot(t, g1, t, g2); plt.legend(("exitatory,solo", "inhibitory, paired", "excitatory, paired")) plt.xlabel('Time(ms)'); plt.ylabel('g_syn(mS/cm^2)'); plt.show() plt.plot(t, V1, t, V2); plt.legend(("excitatory only", "inhibitory and excitatory")) plt.xlabel('Time(ms)'); plt.ylabel('V(mV)'); plt.show() if __name__ == "__main__": main()
Implementation of quadratic integrate-and-fire model with Runge-Kutta method
#-*- coding:utf-8 -*- import numpy as np import matplotlib.pyplot as plt #quadratic integrate and fire model class IzhNeuron: def __init__(self, label, a, b, c, d, v0, u0=None): self.label = label self.a = a; self.b = b self.c = c; self.d = d self.v = v0; self.u = u0 if u0 is not None else b*v0 class IzhSim: def __init__(self, n, T, dt=0.005): self.neuron = n; self.dt = dt self.t = t = np.arange(0, T+dt, dt); self.stim = np.zeros(len(t)) self.x = 5.; self.y = 140. self.du = lambda a, b, v, u: a*(b*v - u) def izhikevich(self, x, t, s, a, b): return np.array([0.04 * x[0]**2 + 5.0*x[0] + 140 - x[1] + s, a*( b*x[0] -x[1] ) ]) def integrate(self, n=None, ng = None): if n is None: n = self.neuron trace = np.zeros((2,len(self.t))) #4 order Runge_Kutta method for i, j in enumerate(self.stim): X = np.array([n.v, n.u]) w1 = self.dt *ng(X, self.t[i], self.stim[i], n.a, n.b) w2 = self.dt * ng(X + w1 * 0.5, self.t[i] * self.dt, self.stim[i], n.a, n.b) w3 = self.dt * ng(X + w2 * 0.5, self.t[i] * self.dt, self.stim[i], n.a, n.b) w4 = self.dt * ng(X + w3, self.t[i] + self.dt, self.stim[i], n.a, n.b) n.v += 1./6. * (w1[0] + 2*w2[0] + 2*w3[0] + w4[0]) n.u += 1./6. * (w1[1] + 2*w2[1] + 2*w3[1] + w4[1]) if n.v > 30: trace[0,i] = 30 n.v= n.c n.u += n.d else: trace[0,i] = n.v trace[1,i] = n.u return trace def main(): sims = [] ## (A) phasic spiking n = IzhNeuron("(A) 5-HT phasic neuron", a=0.005, b=0.28, c=-57., d=2., v0=-60) s = IzhSim(n, T=200) for i, t in enumerate(s.t): s.stim[i] = 1.0 if t > 10 else 0 sims.append(s) ##(B) phasic spiking n = IzhNeuron("(B) GABA phasic neuron", a=0.02,b=0.25,c=-67.,d=2.,v0=-60) s = IzhSim(n, T=200) for i, t in enumerate(s.t): s.stim[i] = 1.0 if t > 10 else 0 sims.append(s) for i,s in enumerate(sims): res = s.integrate(ng=s.izhikevich) plt.plot(s.t, res[0], s.t, -95 + ((s.stim - min(s.stim))/(max(s.stim) - min(s.stim)))*10) plt.show() if __name__ == "__main__": main()
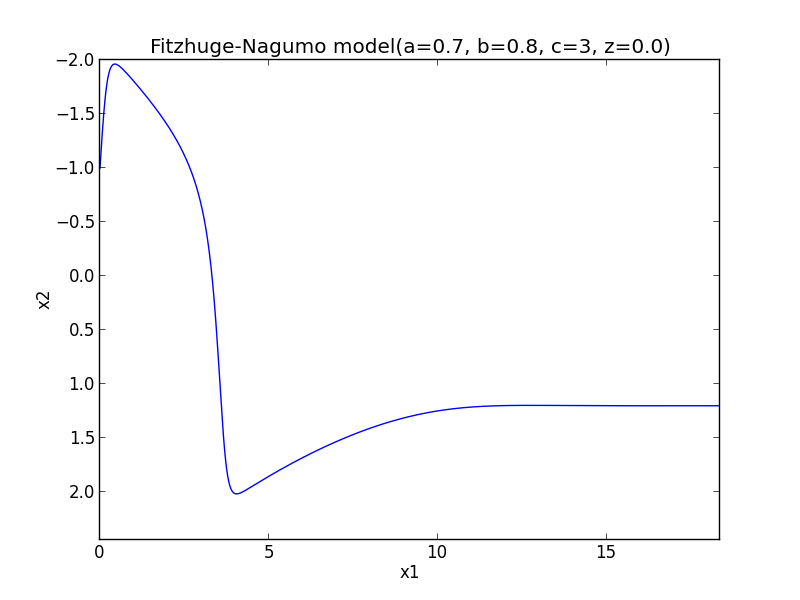
Implementation of Fitzhug-Nagumo model with Runge-Kutta method
#-*- coding:utf-8 -*- import numpy as np import matplotlib.pylab as plt #node def phase_func(x, t, a=-1., b=0., c=0., d=-2.): return np.array([a*x[0]+b*x[1], c*x[0] + d*x[1]]) #focus def phase_focus(x, t, a=1., b=2., c=-2., d=1.): return np.array([a*x[0]+b*x[1], c*x[0] + d*x[1]]) #saddle def phase_saddle(x, t, a=-1., b=-1., c=4., d=1.): return np.array([a*x[0]+b*x[1], c*x[0] + d*x[1]]) #Fitz_Nagumo def fitz_nagumo(x, t, a=0.7, b=0.8, c=3, z=-0.00): return np.array([c*(x[1] + x[0] - x[0]**3/3. + z), -1./c * (x[0] -a + b*x[1])]) def runge_kutta(t0 = 0.0, x0 = np.array([1.0]), t1 = 100., h = 0.01, ng = None): T = np.arange(t0, t1, h) N = np.size(T) X = np.zeros((N, np.size(x0))) X[0] = x0 for i in xrange(1, N): x = X[i-1]; t = T[i-1] w1 = h * ng(x, t) w2 = h * ng(x + w1 * 0.5, t + 0.5 * h) w3 = h * ng(x + w2 * 0.5, t + 0.5 * h) w4 = h * ng(x + w3, t + h) X[i] = X[i-1] + 1./6. * (w1 + 2*w2 + 2*w3 + w4) return X,T def main(): plt.figure() #Node #for i in xrange(-5, 5): # for j in xrange(-5, 5): # X = runge_kutta(x0 = np.array([i, j]), ng = phase_func) # plt.plot(X[:, 0], X[:, 1]) #plt.xlabel('x1') #plt.ylabel('x2') #plt.title('Phase Plane(node) p^2 >4q, q >0') #plt.show() #focus #for i in xrange(-5, 5) : # for j in xrange(-5, 5): # X = runge_kutta(x0 = np.array([i, j]), ng = phase_focus) # plt.plot(X[:, 0], X[:, 1]) #plt.xlabel('x1') #plt.ylabel('x2') #plt.title('Phase Plane(focus) p^2 < 4q, q >0') #plt.show() #saddle #for i in xrange(-5, 5) : # for j in xrange(-5, 5): # X = runge_kutta(x0 = np.array([i, j]), ng = phase_saddle) # plt.plot(X[:, 0], X[:, 1]) #plt.xlabel('x1') #plt.ylabel('x2') #plt.title('Phase Plane(saddle) q < 0') #plt.show() #Fit_Nagumo_model #for i in xrange(-5, 5): # for j in xrange(-5, 5): X, T = runge_kutta(x0 = np.array([-1., -1.]), ng = fitz_nagumo) # plt.plot(X[:, 0], X[:, 1]); plt.plot(T, X[:, 0]) plt.xlabel('x1') plt.ylabel('x2') ax = plt.gca() ax.invert_yaxis() plt.title('Fitzhuge-Nagumo model(a=0.7, b=0.8, c=3, z=0.0)') plt.show() if __name__ == "__main__": main()